Plugin af-dist
This plugin allows to detect possible strand issues by checking genotype frequencies against population allele frequencies.
If working with human data, first download the 1000 Genomes allele frequency annotations
wget -O af.vcf.gz ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/ALL.wgs.phase3_shapeit2_mvncall_integrated_v5b.20130502.sites.vcf.gz bcftools index af.vcf.gz
Then annotate your data file and stream the result through the af-dist plugin to create
the genotype frequency distribution
bcftools annotate -c INFO/AF -a af.vcf.gz data.vcf.gz | bcftools +af-dist | grep ^PROB > data.dist.txt
The output should something like this
PROB_DIST 0.000000 0.100000 100618 PROB_DIST 0.100000 0.200000 144103 PROB_DIST 0.200000 0.300000 214923 PROB_DIST 0.300000 0.400000 320721 PROB_DIST 0.400000 0.500000 817965 PROB_DIST 0.500000 0.600000 84027 PROB_DIST 0.600000 0.700000 86531 PROB_DIST 0.700000 0.800000 97986 PROB_DIST 0.800000 0.900000 108776 PROB_DIST 0.900000 1.000000 176755
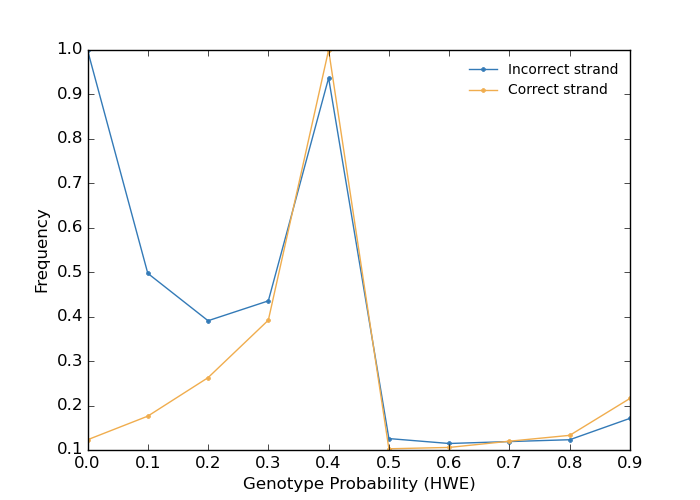
Finally plot the distribution to check whether there are only few unlikely genotypes.
The method is reliable and robust even for non-European populations, as shown below.
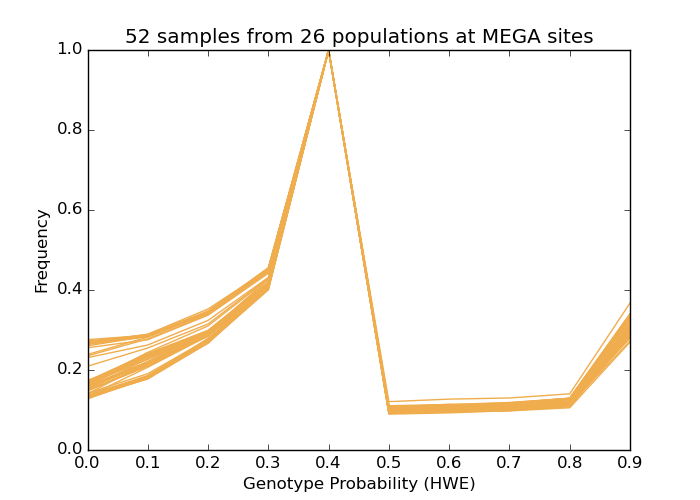
The plot shows af-dist results for 52 samples from 26 populations
of the 1000 Genomes Project (two samples randomly selected from each population), subset to
MEGA
sites, an array with population-specific variants.
Feedback
We welcome your feedback, please help us improve this page by either opening an issue on github or editing it directly and sending a pull request.