Htsget retrieval API spec v1.3.0
This specification describes the htsget protocol version 1.3.0. This printing is version e821e4f from the hts-specs repository.
- Design principles
- Protocol essentials
- GET request
- POST request
- Response
- GA4GH service-info
- GA4GH Service Registry
- Version history
Design principles
This data retrieval API bridges from existing genomics bulk data transfers to a client/server model with the following features:
- Incumbent data formats (BAM, CRAM) are preferred initially, with a future path to others.
- Multiple server implementations are supported, including those that do format transcoding on the fly, and those that return essentially unaltered filesystem data.
- Multiple use cases are supported, including access to small subsets of genomic data (e.g. for browsing a given region) and to full genomes (e.g. for calling variants).
- Clients can provide hints of the information to be retrieved; servers can respond with more information than requested but not less.
- We use the following conventions:
- 0 start, half open coordinates
- JSON format for signaling messages (redirect responses & POST request bodies), describable by OpenAPI Schemas
Explicitly this API does NOT:
- Provide a way to discover the identifiers for valid ReadGroupSets — clients obtain these via some out of band mechanism
This protocol specification is accompanied by a corresponding OpenAPI description. OpenAPI is a language-independent way of describing REST services and is compatible with a number of third party tools. (Note: if there are differences between this text and the OpenAPI description, this specification text is definitive.)
Protocol essentials
All API invocations are made to a configurable HTTP(S) endpoint, receive either URL-encoded query string parameters (GET) or a JSON request body (POST), and return JSON output. Successful requests result with HTTP status code 200 and have UTF8-encoded JSON in the response body. The server may provide responses with chunked transfer encoding. The client and server may mutually negotiate HTTP/2 upgrade using the standard mechanism.
The JSON response is an object with the single key htsget as described in the Response JSON fields and Error Response JSON fields sections. This ensures that, apart from whitespace differences, the message always starts with the same prefix. The presence of this prefix can be used as part of a client’s response validation.
Any timestamps that appear in the response from an API method are given as ISO 8601 date/time format.
HTTP responses may be compressed using RFC 2616 transfer-coding, not content-coding.
Requests adhering to this specification MAY include an Accept header specifying the htsget protocol version they are using:
Accept: application/vnd.ga4gh.htsget.v1.0.0+json
JSON responses SHOULD include a Content-Type header describing the htsget protocol version defining the JSON schema used in the response, e.g.,
Content-Type: application/vnd.ga4gh.htsget.v1.0.0+json; charset=utf-8
Errors
The server MUST respond with an appropriate HTTP status code (4xx or 5xx) when an error condition is detected. In the case of transient server errors, (e.g., 503 and other 5xx status codes), the client SHOULD implement appropriate retry logic as discussed in Reliability & performance considerations below.
For errors that are specific to the htsget protocol, the response body SHOULD be a JSON object (content-type application/json) providing machine-readable information about the nature of the error, along with a human-readable description. The structure of this JSON object is described as follows.
Error Response JSON fields
|
|
Container for response object.
|
The following errors types are defined:
| Error type | HTTP status code | Description |
|---|---|---|
| InvalidAuthentication | 401 | Authorization provided is invalid |
| PermissionDenied | 403 | Authorization is required to access the resource |
| NotFound | 404 | The resource requested was not found |
| PayloadTooLarge | 413 | POST request size is too large |
| UnsupportedFormat | 400 | The requested file format is not supported by the server |
| InvalidInput | 400 | The request parameters do not adhere to the specification |
| InvalidRange | 400 | The requested range cannot be satisfied |
The error type SHOULD be chosen from this table and be accompanied by the specified HTTP status code. An example of a valid JSON error response is:
{
"htsget" : {
"error": "NotFound",
"message": "No such accession 'ENS16232164'"
}
}
Security
The htsget API enables the retrieval of potentially sensitive genomic data by means of a client/server model. Effective security measures are essential to protect the integrity and confidentiality of these data.
Sensitive information transmitted on public networks, such as access tokens and human genomic data, MUST be protected using Transport Level Security (TLS) version 1.2 or later, as specified in RFC 5246.
If the data holder requires client authentication and/or authorization, then the client’s HTTPS API request MUST present an OAuth 2.0 bearer access token as specified in RFC 6750, in the Authorization request header field with the Bearer authentication scheme:
Authorization: Bearer [access_token]
The policies and processes used to perform user authentication and authorization, and the means through which access tokens are issued, are beyond the scope of this API specification. GA4GH recommends the use of the OAuth 2.0 framework (RFC 6749) for authentication and authorization.
CORS
All API resources should have the following support for cross-origin resource sharing (CORS) to support browser-based clients:
If a request to the URL of an API method includes the Origin header, its contents will be propagated into the Access-Control-Allow-Origin header of the response. Preflight requests (OPTIONS requests to the URL of an API method, with appropriate extra headers as defined in the CORS specification) will be accepted if the value of the Access-Control-Request-Method header is GET.
The values of Origin and Access-Control-Request-Headers (if any) of the request will be propagated to Access-Control-Allow-Origin and Access-Control-Allow-Headers respectively in the preflight response.
The Access-Control-Max-Age of the preflight response is set to the equivalent of 30 days.
GET request
Methods
The recommended endpoints to access reads and variants data are:
GET /reads/<id>
GET /variants/<id>
The JSON response is a “ticket” allowing the caller to obtain the desired data in the specified format, which may involve follow-on requests to other endpoints, as detailed below.
The client can request only records overlapping a given genomic range. The response may however contain a superset of the desired results, including all records overlapping the range, and potentially other records not overlapping the range; the client should filter out such extraneous records if necessary. Successful requests with empty result sets still produce a valid response in the requested format (e.g. including header and EOF marker).
The client can also request only read or variant headers. For example, a header request can be used to discover the list of referenceName values applicable for a data stream, provided that the data stream contains @SQ (reads) or ##contig (variants) headers.
URL parameters
|
|
A string identifying which records to return. The format of this identifier is left to the discretion of the API provider, including allowing embedded “/” characters. The following would be valid identifiers:
The id |
Query parameters
|
|
Request data in this format. The allowed values for each type of record are:
The server SHOULD reply with an |
||
|
|
Request different classes of data.
By default, i.e., when If
|
||
|
|
The reference sequence name, for example “chr1”, “1”, or “chrX”. If unspecified, all records are returned regardless of position. For the reads endpoint: if “*”, unplaced unmapped reads are returned. If unspecified, all reads (mapped and unmapped) are returned. (Unplaced unmapped reads are the subset of unmapped reads completely without location information, i.e., with RNAME and POS field values of “*” and 0 respectively. See the SAM specification for details of placed and unplaced unmapped reads.) The server SHOULD reply with a |
||
|
|
The start position of the range on the reference, 0-based, inclusive. The server SHOULD respond with an The server SHOULD respond with an |
||
|
|
The end position of the range on the reference, 0-based exclusive. The server SHOULD respond with an The server SHOULD respond with an |
||
|
|
A list of fields to include, see below. Default: all |
||
|
|
A comma separated list of tags to include, default: all. If the empty string is specified (tags=) no tags are included. The server SHOULD respond with an |
||
|
|
A comma separated list of tags to exclude, default: none. The server SHOULD respond with an |
Field filtering
The list of fields is based on BAM fields:
| Field | Description |
|---|---|
| QNAME | Read names |
| FLAG | Read bit flags |
| RNAME | Reference sequence name |
| POS | Alignment position |
| MAPQ | Mapping quality score |
| CIGAR | CIGAR string |
| RNEXT | Reference sequence name of the next fragment template |
| PNEXT | Alignment position of the next fragment in the template |
| TLEN | Inferred template size |
| SEQ | Read bases |
| QUAL | Base quality scores |
Example: fields=QNAME,FLAG,POS.
As with range filtering, a server may include more fields than explicitly requested, a possibility which clients must be prepared to handle. The fieldsParameterEffective attribute in the GA4GH service-info metadata (see below) indicates whether a server supports filtering individual fields. In any case, servers must not generate any response payload that is invalid under the respective format specification (for example, if a request for reads in BAM format includes QUAL, then the response must provide SEQ as well).
POST request
In addition to the GET method, servers may optionally also accept POST requests. The main differences to the GET method are that query parameters are encoded in JSON format and it is possible to request data for more than one genomic range. The data returned is the same JSON “ticket” as that returned by the GET method, as detailed below.
Htsget POST requests must be “safe” as defined in section 4.2.1 of RFC 7231, that is they must be essentially read-only.
It should be expected that htsget servers will have a limit on the size of POST request that they can accept.
If an incoming POST request is too large, the server SHOULD reply with a PayloadTooLarge error.
URL parameters
POST and GET requests should use the same URLs.
POST requests MUST NOT use any of the query parameters defined for GET requests on the URL.
If query parameters are present, the server SHOULD reply with an InvalidInput error.
Request body
The POST request body is a JSON object containing the request parameters,
which have similar names and meanings to those used in the GET query.
A notable difference is that the referenceName, start and end parameters are not used at the top level of the JSON object.
Instead as they are put into an array under the regions key, which allows more than one range to be specified.
|
|
Request data in this format. The allowed values for each type of record are:
The server SHOULD reply with an |
||||||
|
|
Request different classes of data.
As for GET requests, if
|
||||||
|
|
List of fields to include. See Field filtering.
|
||||||
|
|
List of tags to include, default: all. If an empty array is specified, no tags are included. The server SHOULD respond with an
|
||||||
|
|
List of tags to exclude, default: none. The server SHOULD respond with an
|
||||||
|
|
Regions to return. If not present, the entire file will be returned. When present, the array must contain at least one region specification. Note that regions will be returned in the order that they appear in the file, which may not match the order in the list. Any overlapping regions will be merged. The server SHOULD respond with an
|
Example
{
"format" : "bam",
"fields" : ["QNAME", "FLAG", "RNAME", "POS", "CIGAR", "SEQ"],
"tags" : ["RG"],
"notags" : ["OQ"],
"regions" : [
{ "referenceName" : "chr1" },
{ "referenceName" : "chr2", "start" : 999, "end" : 1000 },
{ "referenceName" : "chr2", "start" : 2000, "end" : 2100 }
]
}
Region list
The regions parameter is an array of objects which describe the locations to be returned.
Each location object contains a referenceName, which must always be present, and optional start and end tags.
The first region in the example above shows how to return all reads for reference “chr1”.
To return reads covering a single base, the client should use an end position one greater than the start, as shown in the second region in the example which requests the 1000th base of reference “chr2”.
The regions list acts as a filter on the requested file. Records that overlap the requested regions will be returned in the order that they occurred in the original file. This may not be the same as the order in the list. A record will only be returned once, even if it matches more than one location in the list.
As with the GET request, the server response may contain a super-set of the desired results. Clients will need to filter out any extraneous records if necessary.
Response
Response JSON fields
|
|
Container for response object.
|
An example of a JSON response is:
{
"htsget" : {
"format" : "BAM",
"urls" : [
{
"url" : "data:application/vnd.ga4gh.bam;base64,QkFNAQ==",
"class" : "header"
},
{
"url" : "https://htsget.blocksrv.example/sample1234/header",
"class" : "header"
},
{
"url" : "https://htsget.blocksrv.example/sample1234/run1.bam",
"headers" : {
"Authorization" : "Bearer xxxx",
"Range" : "bytes=65536-1003750"
},
"class" : "body"
},
{
"url" : "https://htsget.blocksrv.example/sample1234/run1.bam",
"headers" : {
"Authorization" : "Bearer xxxx",
"Range" : "bytes=2744831-9375732"
},
"class" : "body"
}
]
}
}
Response data blocks
Diagram of core mechanic
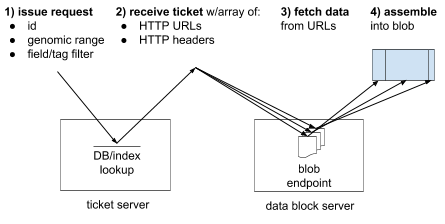
- Client sends a request with id, genomic range, and filter.
- Server replies with a ticket describing data block locations (URLs and headers).
- Client fetches the data blocks using the URLs and headers.
- Client concatenates data blocks to produce local blob.
While the blocks must be finally concatenated in the given order, the client may fetch them in parallel and/or reuse cached data from URLs that have previously been downloaded.
When making a series of requests to fetch reads or variants within different regions of the same <id> resource, clients may wish to avoid re-fetching the SAM/CRAM/VCF headers each time, especially if they are large.
If the ticket contains class fields, the client may reuse previously downloaded and parsed headers rather than re-fetching the header-class URLs.
HTTPS data block URLs
- must have percent-encoded path and query (e.g. JavaScript
encodeURIComponent; Pythonurllib.urlencode) - must accept GET requests
- should provide CORS
- should allow multiple request retries, within reason
- should use HTTPS rather than plain HTTP except for testing or internal-only purposes (providing both security and robustness to data corruption in flight)
- need not use the same authentication scheme as the API server. URL and
headersmust include any temporary credentials necessary to access the data block. Client must not send the bearer token used for the API, if any, to the data block endpoint, unless copied in the requiredheaders. - Server must send the response with either the Content-Length header, or chunked transfer encoding, or both. Clients must detect premature response truncation.
- Client and URL endpoint may mutually negotiate HTTP/2 upgrade using the standard mechanism.
- Client must follow 3xx redirects from the URL, subject to typical fail-safe mechanisms (e.g. maximum number of redirects), always supplying the
headers, if any. - If a byte range HTTP header accompanies the URL, then the client MAY decompose this byte range into several sub-ranges and open multiple parallel, retryable requests to fetch them. (The URL and
headersmust be sufficient to authorize such behavior by the client, within reason.)
Inline data block URIs
e.g. data:application/vnd.ga4gh.bam;base64,SGVsbG8sIFdvcmxkIQ== (RFC 2397, Data URI).
The client obtains the data block by decoding the embedded base64 payload.
- must use base64 payload encoding (simplifies client decoding logic)
- client should ignore the media type (if any), treating the payload as a partial blob.
Note: the base64 text should not be additionally percent encoded.
Reliability & performance considerations
To provide robustness to sporadic transfer failures, servers should divide large payloads into multiple data blocks in the urls array. Then if the transfer of any one block fails, the client can retry that block and carry on, instead of starting all over. Clients may also fetch blocks in parallel, which can improve throughput.
Initial guidelines, which we expect to revise in light of future experience:
- Data blocks should not exceed ~1GB
- Inline data URIs should not exceed a few megabytes
Security considerations
The data block URL and headers might contain embedded authentication tokens; therefore, production clients and servers should not unnecessarily print them to console, write them to logs, embed them in error messages, etc.
GA4GH service-info
Following the GA4GH service-info specification, htsget servers SHOULD also expose /reads/service-info and/or /variants/service-info metadata endpoints. In addition to the standard service-info fields, including {"type": {"group": "org.ga4gh", "artifact": "htsget", "version": "x.y.z"}}, the response SHOULD include an htsget object further describing htsget-specific capabilities, with the following fields:
|
|
Indicates the htsget datatype category (‘reads’ or ‘variants’) served by the ticket endpoint related to this service-info endpoint. Use either:
|
|
|
List of requested
|
|
|
Indicates whether the service is capable of returning only a subset of available fields based on the |
|
|
Indicates whether the service is capable of server-side result tag inclusion/exclusion using the |
Example service-info response:
{
"id": "net.exampleco.htsget",
"name": "ExampleCo genomics data service",
"version": "0.1.0",
"organization": {
"name": "ExampleCo",
"url": "https://exampleco.com"
},
"type": {
"group": "org.ga4gh",
"artifact": "htsget",
"version": "1.3.0"
},
"htsget": {
"datatype": "reads",
"formats": ["BAM", "CRAM"],
"fieldsParameterEffective": true,
"tagsParametersEffective": false
}
}
GA4GH Service Registry
The GA4GH Service Registry API specification allows information about GA4GH-compliant web services, including htsget services, to be aggregated into registries and made available via a standard API. The following considerations SHOULD be followed when registering htsget services within a service registry.
- Endpoints for different htsget data types should be registered as separate entities within the registry. If an htsget service provides both
readsandvariantsdata, both endpoints should be registered. - The
urlproperty should reference the API’s ticket endpoint for a single data type, excluding any particularid(i.e. an htsget reads API registration should provide the URL to the reads ticket endpoint, an htsget variants API registration should provide the URL to the variants ticket endpoint). Clients should be able to assume that:- Adding
/{id}to the registeredurlwill hit the corresponding ticket endpoint (i.e./reads/{id}or/variants/{id}). - Adding
/service-infoto the registeredurlwill hit the correspondingservice-infoendpoint (i.e./reads/service-infoor/variants/service-info).
- Adding
- The value of the
typeobject’sartifactproperty should behtsget(i.e. the same as it appears inservice-info)
Example listing of htsget reads API and variants API registrations from a service registry’s /services endpoint:
[
{
"id": "net.exampleco.htsget-reads",
"name": "ExampleCo htsget reads API",
"description": "Serves alignment data (BAM, CRAM) via htsget protocol",
"version": "0.1.0",
"url": "https://htsget.exampleco.com/v1/reads",
"organization": {
"name": "ExampleCo",
"url": "https://exampleco.com"
},
"type": {
"group": "org.ga4gh",
"artifact": "htsget",
"version": "1.3.0"
}
},
{
"id": "net.exampleco.htsget-variants",
"name": "ExampleCo htsget variants API",
"description": "Serves variant data (VCF, BCF) via htsget protocol",
"version": "0.1.0",
"url": "https://htsget.exampleco.com/v1/variants"
"organization": {
"name": "ExampleCo",
"url": "https://exampleco.com"
},
"type": {
"group": "org.ga4gh",
"artifact": "htsget",
"version": "1.3.0"
}
}
]
Version history
This appendix lists the significant functionality introduced and changes made in each published version of the htsget protocol.
1.3.0 (March 2021)
Added POST requests for both reads and variants endpoints, allowing data to be queried via the HTTP POST method, and defining a JSON request object to be used as the POST payload instead of the corresponding set of htsget query parameters.
Defined a new PayloadTooLarge error type.
Added a service-info endpoint and defined an htsget service-info response sub-object with datatype, formats, fieldsParameterEffective, and tagsParametersEffective optional fields.
Added discussion of registering htsget endpoints in a GA4GH service registry.
1.2.0 (May 2019)
Added a class=header query parameter for requesting headers only rather than including reads/variants data records.
Defined response ticket "class": "header"|"body" optional field to enable reusing previously downloaded URL data when making repeated requests to the same <id> resource.
1.1.1 (January 2019)
Added referenceName=* special case for requesting unplaced unmapped reads from a reads endpoint.
1.1.0 (June 2018)
Added the GET /variants/<id> endpoint, allowing for querying VCF/BCF variants data.
1.0.0 (October 2017)
The first published version of the htsget protocol, which specified only the GET /reads/<id> endpoint.
BAM/CRAM sequencing data could be queried via the HTTP GET method using format, referenceName/start/end, fields, and tags/notags query parameters.